In the rapidly evolving field of materials science, researchers have long faced a significant challenge: efficiently communicating complex polymer structures. The development of a compact yet robust notation system for molecular structures has become crucial for advancing AI-driven polymer research and facilitating seamless knowledge exchange within the scientific community. While small molecules have enjoyed standardized representation methods, the polymer sector has struggled with creating an efficient system—until now.
Unlike small molecules with well-defined structures, polymers present unique challenges due to their inherently stochastic nature. These complex macromolecules exist as ensembles with diverse structural distributions, rendering traditional deterministic representations inadequate. In a groundbreaking study published on September 12 in ACS Central Science, collaborative researchers from MIT, Duke University, and Northwestern University unveiled BigSMILES—an innovative notation system specifically designed to handle the randomness characteristic of polymer structures.
"BigSMILES represents a quantum leap in digital polymer representation," explains Connor Coley PhD '19, co-author of the paper. "Unlike small molecules, polymers typically exist as multiple structural ensembles created through stochastic processes. This fundamental difference necessitated an entirely new approach to structural notation—one that BigSMILES now provides."
The research team comprises distinguished scientists including Coley; MIT's associate professor of chemical engineering Bradley D. Olsen; Klavs F. Jensen, Warren K. Lewis Professor of Chemical Engineering at MIT; Northwestern University's assistant professor of chemistry Julia A. Kalow; MIT's associate professor of chemistry Jeremiah A. Johnson; Duke University's Stephen L. Craig, William T. Miller Professor of Chemistry; along with graduate students Eliot Woods from Northwestern, Zi Wang from Duke, Wencong Wang, Haley K. Beech, Tzyy-Shyang Lin, and visiting researcher Hidenobu Mochigase from MIT.
While several line notations exist for molecular structure communication, the simplified molecular-input line-entry system (SMILES) has emerged as the predominant choice. SMILES offers exceptional human readability and extensive software support, making it ideal for labeling chemical data and facilitating compact information exchange between researchers. Its text-based nature also makes it particularly compatible with AI and machine learning algorithms, enabling successful applications in small molecule property prediction and computer-aided synthesis planning.
However, polymers have stubbornly resisted description by SMILES and similar structural languages. Most existing systems were designed to represent well-defined atomistic graphs, a paradigm that doesn't align with the stochastic nature of polymers. This lack of unified polymer representation has significantly hindered the advancement of polymer informatics. While pioneering initiatives like the Polymer Genome Project have demonstrated the potential of SMILES extensions, the rapid evolution of new chemistry and materials informatics has underscored the urgent need for a universally applicable polymer notation system.
"Machine learning offers tremendous potential to accelerate chemical discovery and development," notes Lin He, acting deputy division director for the National Science Foundation (NSF) Division of Chemistry. "This innovative structural labeling tool, specifically engineered to address polymers' unique challenges, significantly enhances chemical structural data searchability. It brings us closer to fully harnessing the data revolution in materials science."
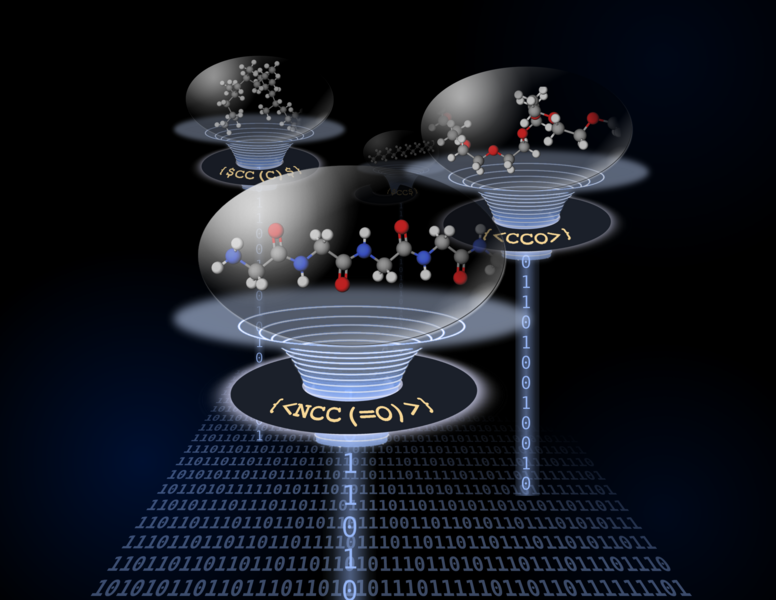
The researchers have developed a groundbreaking structural construct as an extension of the highly successful SMILES representation, specifically designed to accommodate the random nature of polymer materials. Given the high molar mass of these macromolecules, this innovative system has been aptly named BigSMILES. Within this framework, polymeric fragments are represented using repeating units enclosed in curly brackets. The chemical structures of these units employ standard SMILES syntax but incorporate additional bonding descriptors that specify connections between different repeating units. This elegant design enables the encoding of diverse macromolecules across various chemistries, including homopolymers, random copolymers, block copolymers, and structures ranging from linear to ring to branched polymers. Like its predecessor, BigSMILES delivers compact, self-contained text representations.
"Standardizing polymer digital representation through BigSMILES will foster unprecedented data sharing and aggregation in polymer science," observes Jason Clark, materials lead in Open Innovation for Renewable Chemicals and Materials at Braskem, who wasn't involved in the research. "This system represents a significant breakthrough by addressing the critical need for a flexible framework to digitally represent complex polymer structures. As the industry embraces AI-driven research methodologies, BigSMILES will undoubtedly accelerate innovation cycles."
Clark adds, "The plastics industry faces numerous challenges in the circular economy context, from raw material sourcing to end-of-life management. Addressing these issues requires innovative polymer-based material design, traditionally hampered by lengthy development cycles. AI and machine learning have already demonstrated remarkable potential in accelerating development for metal alloys and small organic molecules. BigSMILES digital representations will enable the application of data science methods to evaluate structure-performance relationships, ultimately accelerating the discovery of polymer structures that will advance the circular economy."
"The beauty of BigSMILES lies in its simplicity—complex polymer structures can be constructed using just three new basic operators combined with original SMILES symbols," explains Olsen. "This development impacts numerous fields including polymer science, biomaterials, materials chemistry, and biochemistry—all disciplines built upon macromolecules with stochastic structures. Essentially, we've created a new language for writing the structures of large molecules."
"I'm particularly excited about the potential to directly link data entry with the synthetic methods used to create specific polymers," remarks Craig. "This connection could capture and process more molecular information than typically available through standard characterization techniques. If successfully implemented, it could unlock countless new discoveries in polymer science and materials engineering."
This research was made possible through funding from the NSF via the Center for the Chemistry of Molecularly Optimized Networks, an NSF Center for Chemical Innovation.